Colorectal or bowel cancer originates from either the colon or the rectum. It is the third most common cancer and has the fourth highest mortality rate reported worldwide. The estimated incidence rates of colorectal cancer
are highest in Australia/New Zealand and Western Europe.

Identification of sequence variants in genes has advanced our understanding of how cancer develops, progresses and how these sequence variants can be targeted for a cure. Thanks to the ongoing work of several researchers around the world this is made possible. However, the obtained results are isolated across thousands of scientific literature and websites. A resource that can integrate all these heterogeneous data can aid the biomedical research community in the battle against colorectal cancer.
Colorectal Cancer Atlas is a database that catalogs multiple data types pertaining to
- Quantitative and non-quantitative protein expression data obtained from various techniques including mass spectrometry, Western blotting, immunohistochemistry, confocal microscopy, immunoelectron microscopy and FACS
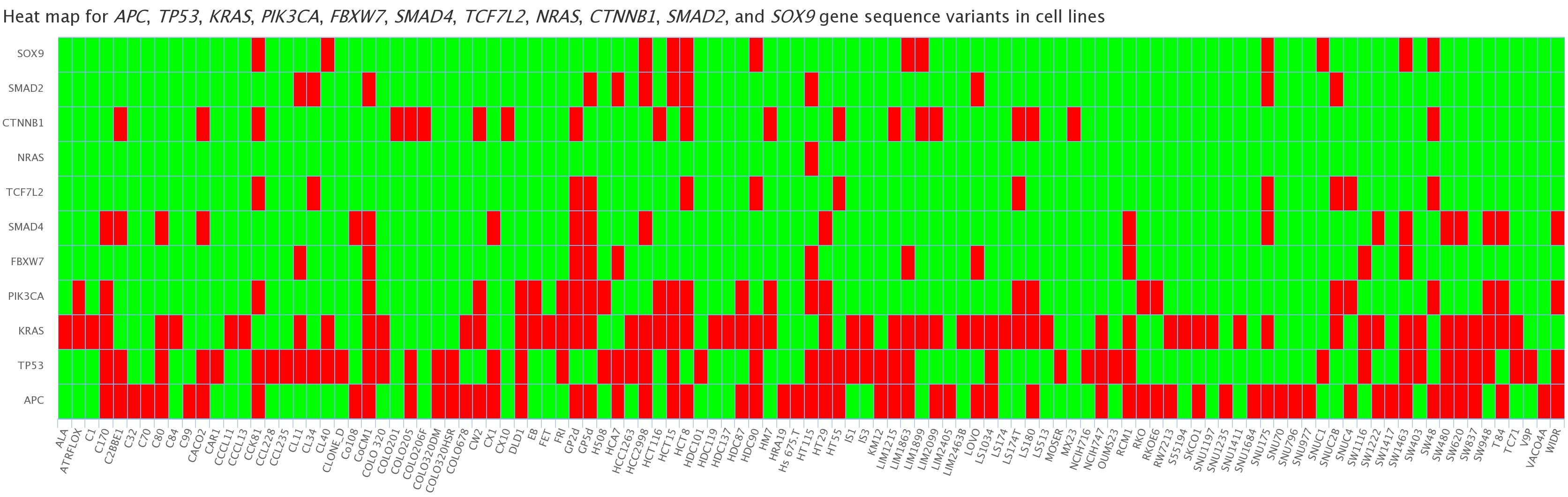
- Mutation data obtained by large and small scale sequencing
- Pathway data from Reactome, NCI, Cell map and HumanCyc
- Protein-protein interactions from BioGRID and HPRD
- Gene Ontology data from Entrez Gene
 Colorectal Cancer Atlas citation
Colorectal Cancer Atlas citation
Chisanga, D., Keerthikumar, S., Pathan, M., Ariyaratne, D., Kalra, H., Boukouris, S., Mathew, N., Al Saffar, H., Gangoda, L., Ang, C.S., Sieber, O., Mariadason, J., Dasgupta, R., Chilamkurti, N. and Mathivanan, S. (2016)
Colorectal Cancer Atlas: An integrative resource for genomic and proteomic annotations from colorectal cancer cell lines and tissue.
Nucleic Acids Research. 44(D1), D969-74
 Quick search
Quick search
- Gene symbol e.g. APC
- Protein name e.g. Adenomatous polyposis coli
- Cell line e.g. HCT116
- Sequence variant e.g. Deletion
- Pathway e.g. Wnt signaling
 Statistics
Statistics
|
Protein identifications
|
|
|
MS/MS spectra
|
|
|
Cell lines
|
|
|
Tissues
|
|
|
Genes with sequence variants
|
|
|
Sequence variants
|
|
|
Pathways with sequence variants
|
|
|
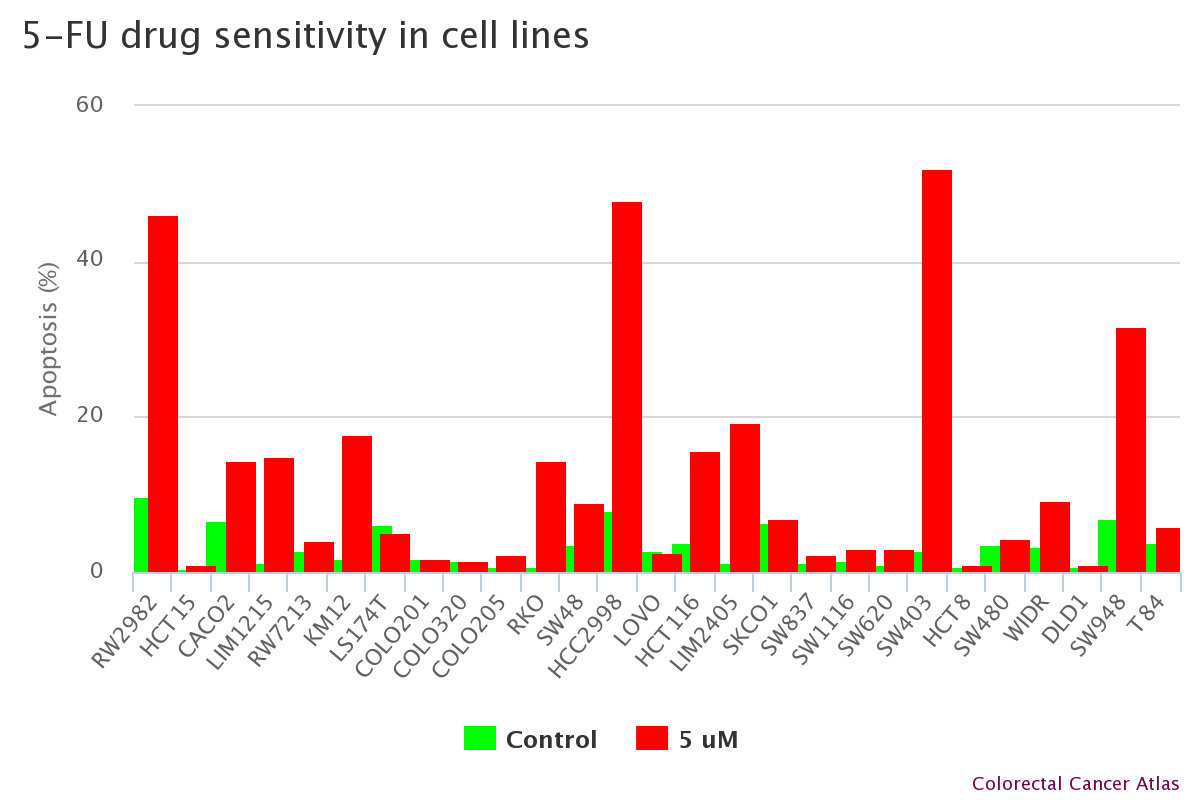
Cell lines with drug sensitivity
|
|
|
PTMs
|
88,819
|
|
PTMs affected by sequence variants
|
|
|
Protein-protein interactions
|
253,700
|
